本片笔记记录我学习用于预测蛋白质特性的机器学习:全面综述 - ScienceDirect 该文章所收集到的有用信息
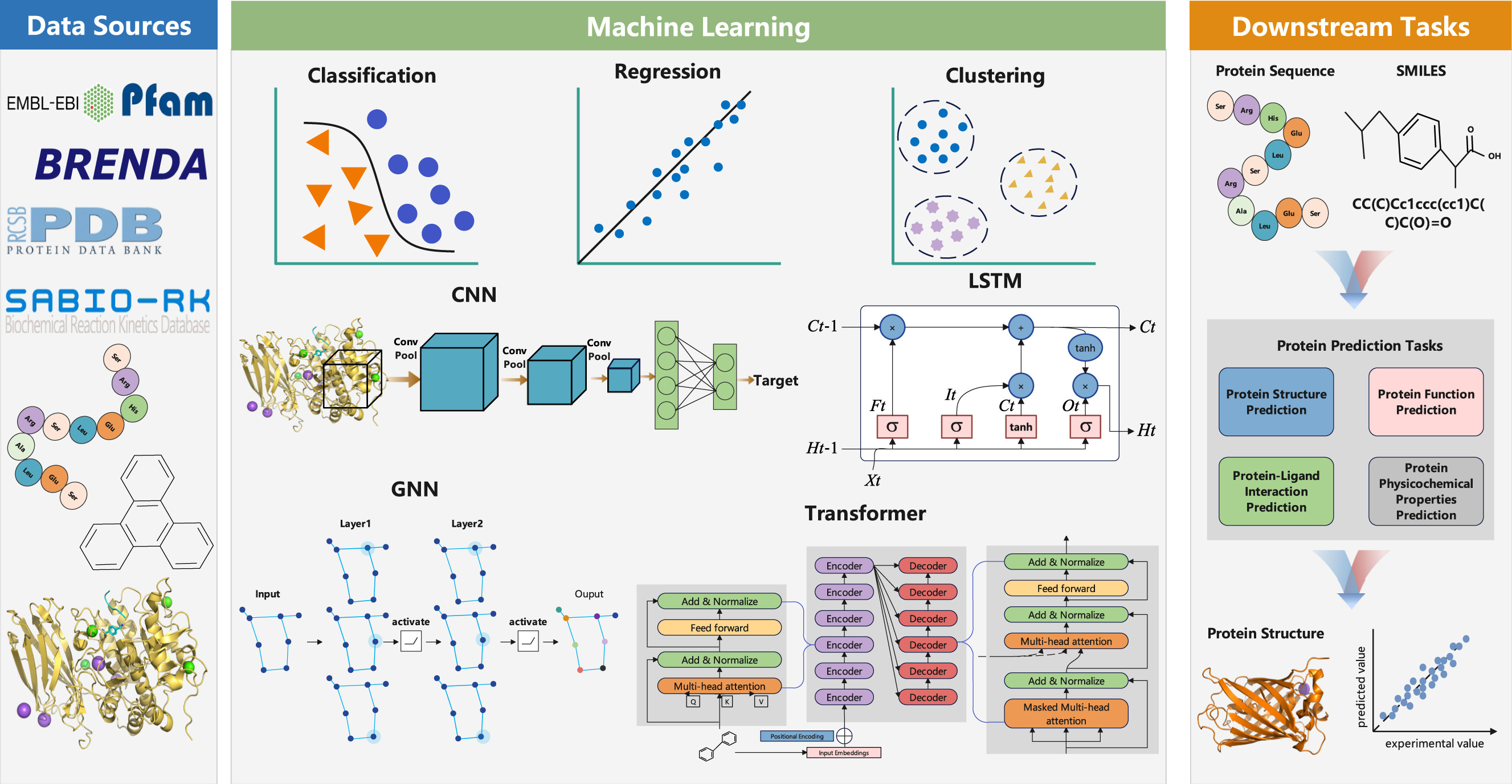
常用的蛋白质序列、结构和功能数据库
| Application | Database | Disk Space | Description | Link |
|---|---|---|---|---|
| Protein Structure | AlphaFoldDB | 23TB | Store 3D structural data of proteins. | https://alphafold.com/ |
| BioLiP | 100GB | A semi-manually curated database for high-quality data. | https://zhanggroup.org/BioLiP/ | |
| ChEMBL | 2TB | A database of bioactive molecules with drug-like properties. | https://www.ebi.ac.uk/chembl/ | |
| PDB | 50TB | 3D structures (protein, nucleic acids). | https://www.rcsb.org/ | |
| PubChem | 300TB | Including chemical and physical characteristics. | https://pubchem.ncbi.nlm.nih.gov/ | |
| STRING | 20TB | 59.3M protein data. | https://string-db.org/ | |
| UniProt | 20TB | Composed of UniParc, UniRef, and UniProtKB. | https://www.uniprot.org/ | |
| ZINC | 17GB | Contains over 35M purchasable compounds. | https://zinc12.docking.org/ | |
| BFD | 270GB | Created by clustering 2.5B protein sequences. | https://bfd.mmsegs.com/ | |
| Protein Function | BRENDA | 63MB | Contains more than 84K proteins and 104K enzyme entries. | https://www.brenda-enzymes.org/ |
| ExPASy ENZYME | 23MB | Describes characteristics of enzymes and metabolic pathways. | https://enzyme.expasy.org/ | |
| GO knowledge-base | 500MB | Includes 4.2K GO terms, 7.6M annotations, 1.5M gene products, and 5.4K species. | https://www.geneontology.org/ | |
| KEGG ENZYME | - | Provides enzyme classification, function, and metabolic channel information. | https://www.genome.jp/kegg/ | |
| PDBbind | 3.5GB | Provides binding affinity data for 23K biomolecular complexes. | http://pdbbind.org.cn/ | |
| SABIO-RK | - | Kinetic data of biochemical reactions. | http://sabio.h-its.org/ | |
| ScOPe | 175MB | Protein structure classification database. | https://scop.berkeley.edu/ | |
| Protein-Ligand Interaction | DUD-E | 2.6GB | 22K active compounds and their affinities against 102 targets. | https://dude.docking.org/ |
| CrossDocked2020 | 90GB | More than 22M ligand-protein affinity scores. | https://bits.csb.pitt.edu/files/crossdock2020/ | |
| BindingDB | 4.45GB | Contains about 2.8M binding data for 9.3K proteins. | https://www.bindingdb.org/ | |
| STITCH | 379.35GB | A resource to explore known and predicted interactions between chemicals. | http://stitch.embl.de/ | |
| Protein Physicochemical Properties | ProTherm | - | Contains experimentally determined thermodynamic parameters of proteins. | https://web.iitm.ac.in/bioinfo2/prothermdb |
| Pfam | 416GB | A collection of protein families, containing 21K entries. | http://pfam-legacy.xfam.org/ | |
| ThermoMutDB | 122MB | Containing over 14K experimental data of thermodynamic properties. | https://biosig.lab.uq.edu.au/thermomutdb/ | |
| Aggrescan3D | 287MB | Provides analysis of solubility and aggregation propensities. | https://biocomp.chem.uw.edu.pl/A3D2/hproteome |
备注:
- 表中的磁盘空间是压缩格式下的估计数据。
- 如果某些字段显示为“-”,表示相关数据可能无法直接获取。
现有的蛋白质和分子预训练模型:
| Application | Classification | Method | Input | Architecture | Database | Year |
|---|---|---|---|---|---|---|
| Protein Structure Prediction | Secondary structure prediction | CFLM [96] | Sequence | RDN | PDB, CASP | 2023 |
| CondGCNN [95] | Sequence | CNN, RNN | CullPDB, CB513, CASP | 2022 | ||
| Tertiary structure prediction | Cerebra [112] | Sequence, MSA | Transformer | PDB, CAMEO | 2024 | |
| Evo [108] | Sequence | StripedHyena | OpenGenome | 2024 | ||
| ESMFold [67] | Sequence | Transformer | UniRef | 2022 | ||
| trRosettaX-Single [102] | Sequence | Transformer, CNN | PDB, Orphan54 | 2022 | ||
| RoseTTAFold [101] | Sequence, Structure, MSA | Transformer | UniProt, BFD | 2021 | ||
| AlphaFold2 [100] | Sequence, Structure, MSA | Transformer | UniProt, PDB, BFD | 2021 | ||
| Protein Function Prediction | GO prediction | Struct2GO [115] | Sequence, Structure | GNN, RNN | EMBL-EBI, GO | 2023 |
| DeepFRI [118] | Sequence, Structure | GNN | GO, PDB, CSA | 2021 | ||
| EC prediction | CLEAN [120] | Sequence | CNN | UniProt | 2023 | |
| HDMLF [121] | Sequence | RNN | UniProt | 2023 | ||
| EC&GO prediction | GearNet [7] | Structure | GNN | EMBL-EBI, UniProt, GO | 2022 | |
| Protein-Ligand Interactions | Affinity prediction | PocketAnchor [122] | Sequence, Structure | GNN | CASF, PDBbind | 2023 |
| GraphscoreDTA [123] | Sequence, Structure | GNN | SIFTS, PDBbind | 2023 | ||
| ProtNet [124] | Sequence, Structure | GNN | PDBbind | 2023 | ||
| Transformer-M [63] | Sequence, Structure | Transformer | PDBbind | 2022 | ||
| 3D-CNNs, SG-CNNs [125] | Sequence, Structure | CNN | PDBbind | 2021 | ||
| DeepAffinity [126] | Sequence | RNN, CNN, GNN | PDBbind, UniRef | 2019 | ||
| Protein Physicochemical Properties Prediction | Stability | RaSP [131] | Sequence, Structure | CNN | ThermoMutDB | 2024 |
| Solubility | PON-Sol2 [134] | Sequence | Random Forest | PON-Sol | 2021 | |
| SoluProt [135] | Sequence | CNN | Target-Track | 2021 | ||
| Subcellular localization | DeepLoc2.0 [136] | Sequence | LSTM | UniProt | 2022 | |
| Binding energy | super-HTS [137] | Sequence | GNN | Created by Rosetta | 2016 |
非冗余蛋白质序列数据库
UniRef 数据集是从 UniProt 和选定的 UniParc 记录中提取的序列簇的集合,旨在通过隐藏冗余序列来覆盖多个分辨率级别的序列空间。该数据集根据 100%、90% 和 50% 序列相似性标准对序列进行聚类,从而通过相似序列的聚类来更快地进行序列比对。BFD 数据库包含超过 2.5B 序列,涵盖细菌、古细菌、真核生物和其他生物界,确保数据多样性并有助于捕获蛋白质功能和结构的广泛变化。UniRef 和 BFD 广泛用于 AlphaFold2 等蛋白质结构预测方法。
非冗余蛋白质结构数据库
一些模型可能会利用相似蛋白质的结构信息来帮助进行结构预测。最大的结构数据库蛋白质数据库 (PDB) 广泛用于收集 3D 蛋白质结构、核酸及其复合物,目前包含近 218K 的实际结构条目。这些数据大多是通过 X 射线晶体学、核磁共振 (NMR) 波谱和冷冻电子显微镜 (cryo-EM) 等实验方法获得的。PDB70 是从 PDB 数据库生成的特定数据集,它使用聚类算法将相似的蛋白质结构组织成簇。每个簇代表一组序列相似度高的蛋白质结构,从而提高模板搜索时的搜索效率。
关于RNN和LSTM
众所周知,RNN 容易出现梯度爆炸和消失的问题,并且在处理长序列数据时难以捕获长期依赖性。为了解决这些问题,引入了长短期记忆网络 (LSTMs) 架构。LSTM 可以看作是 RNN 的整体优化,引入了门控单元和存储机制。门控单元包括忘记门、输入门和输出门,控制信息流并允许 LSTM 选择性地记住或忘记信息。与传统的 RNN 不同,LSTM 在处理序列数据时计算三个门控单元的值,从而决定是忘记、更新还是输出信息。随后,LSTM 根据这些值更新存储单元的内容,从而能够选择性地记住或忘记信息,从而解决了在处理长序列数据时捕获长期关系的问题。一些研究方法对原始 LSTM 进行了改进,主要区别在于门控机制、存储单元和参数共享方法的选择和修改。与 CNN 一样,LSTM 适用于处理蛋白质序列数据,并已用于蛋白质功能预测和蛋白质-配体相互作用预测等任务。
现有蛋白质的研究范式
使用AlphaFold2的结构预测,聚类研究是否存在酶的新功能(一篇Cell工作)
最近,Huang 等人介绍了一种基于结构的蛋白质聚类方法,用于发现脱氨酶功能并鉴定新的脱氨酶家族(Discovery of deaminase functions by structure-based protein clustering: Cell).他们应用 AlphaFold2 来预测蛋白质结构,随后通过结构比对根据预测的结构相似性对整个脱氨酶蛋白家族进行聚类。他们发现了脱氨酶蛋白和新脱氨酶的新功能;这样的发现不能通过挖掘氨基酸序列来获得。

Comments NOTHING